


 |  |  | Reading the output |
The text output simply presents the list of genes predicted with possible partial genes on the border of the sequence and possible frameshifts. For each region predicted, you can read the beginning, the end and the coding phase of the coding region predicted.
Using the first example sequence available on the web, a noisy high GC content sequence with 'N's, selecting the correct organism (Ralstonia) and a frameshift penalty of 8 (for low quality sequences), we get the following prediction:
Start End Ph.
148 911 1 |
| 912 1181 3
1254 1387 3 >
A first gene is predicted on phase 1, from 148 to 1181 interrupted by a predicted frameshift between position 911 and 912. Then a partial gene starts at 1254 on phase 3.
As an illustration, Figure * presents the graphical output obtained using the same GC rich and noisy sequence provided as example 1 on the web interface, using the correct "Ralstonia" model with default parameters and the default frameshift penalty set to 8 (for a low quality sequence).
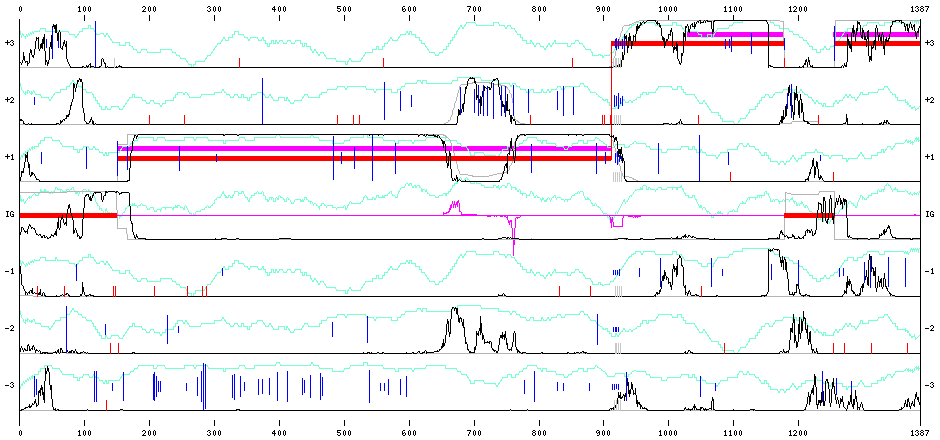
On the above image, one can observe here, between 100 and 150 that FrameD hesitates between the two possible start codons: a non negligible part of the predictions would actually use the second START but the first is preferred. Another position of doubt is in the 700 region where the uncertainty on coding phase is clear: two frameshifts around 650 and 750 roughly could explain the increase in coding score in phase 2. Despite the fact that many suboptimal prediction actually predict these 2 frameshift, the optimal prediction does not.
These two possible frameshifts are also visible on the central magenta line which represent an amplified frameshift expectation based on all possible predictions. We see a possible insertion around 650 (up curve) and a possible deletion around 750 (down curve).
A frameshift is actually predicted around 900, close to the N stretch, close to where we created it.
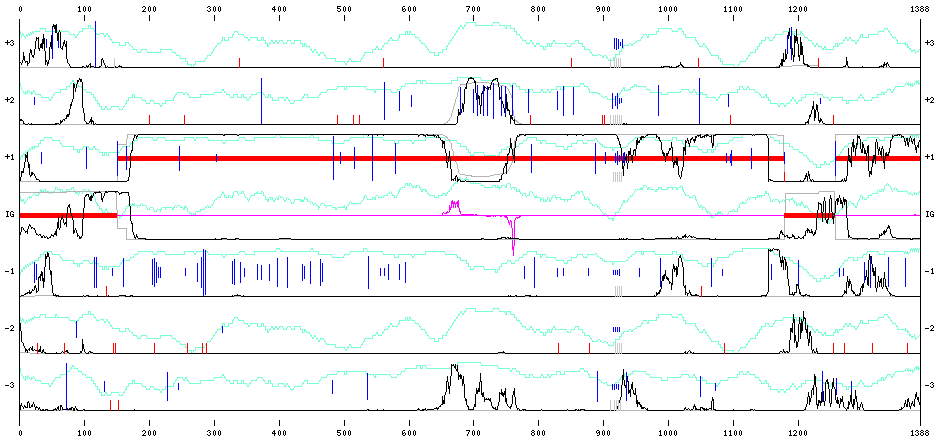
We can get back to the initial page and ask for a corrected sequence and resubmit it for analyze, using the same frameshift penalty of 8 (for low quality). The corresponding graphical output is visible in Figure *.
Start End Ph. 148 1182 1 1255 1388 1 >
No frameshift is predicted anymore.
 |  |  | Reading the output |