![[OK]](Icons/tick.png) Basic Statistics
Basic Statistics
| Measure | Value |
|---|---|
| Filename | s_2_uT22.fastq |
| File type | Conventional base calls |
| Encoding | Illumina 1.5 |
| Total Sequences | 91418250 |
| Filtered Sequences | 0 |
| Sequence length | 36 |
| %GC | 43 |
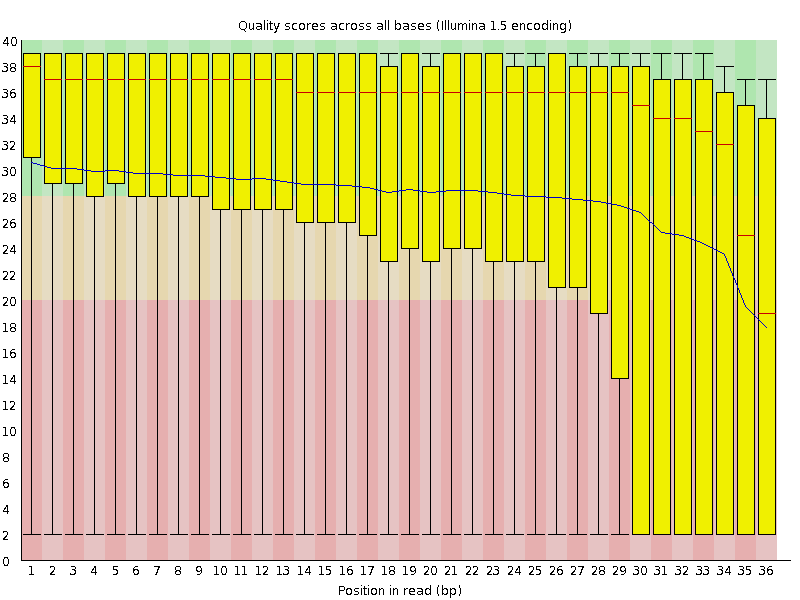
![[FAIL]](Icons/error.png) Per base sequence quality
Per base sequence quality
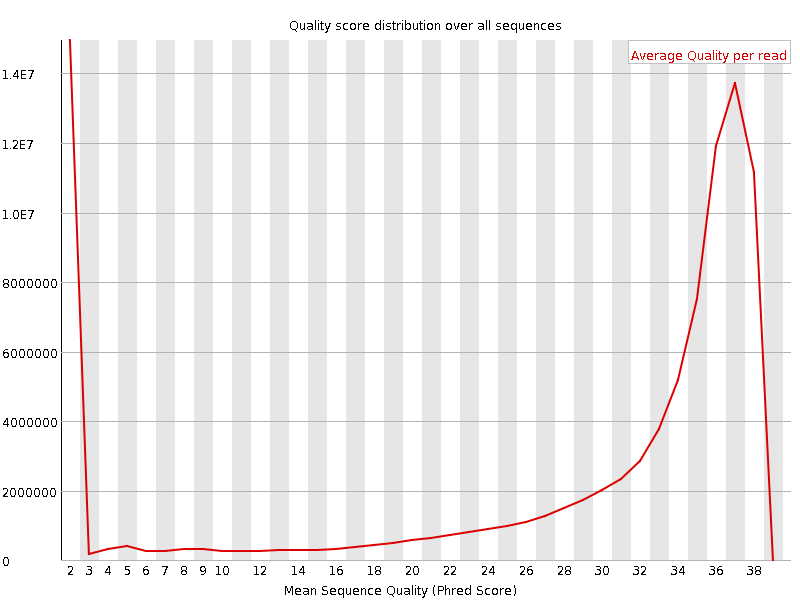
![[FAIL]](Icons/error.png) Per sequence quality scores
Per sequence quality scores
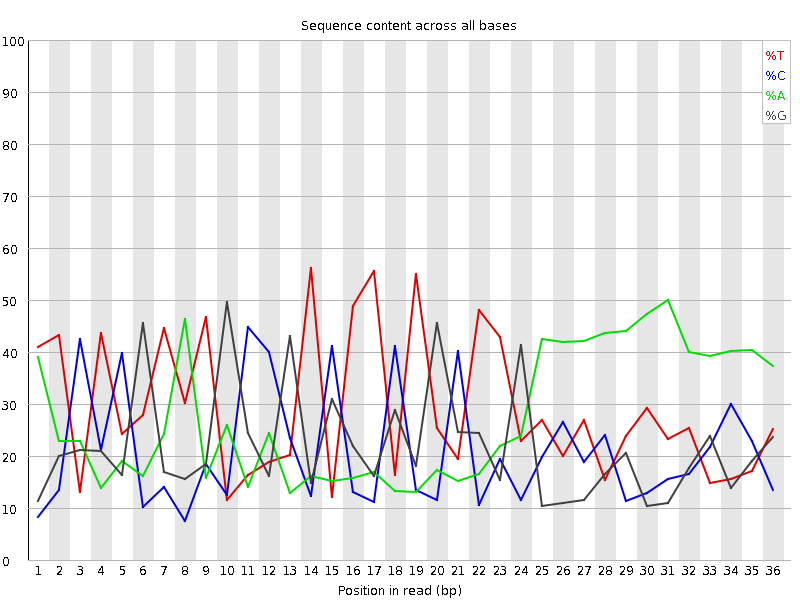
![[FAIL]](Icons/error.png) Per base sequence content
Per base sequence content
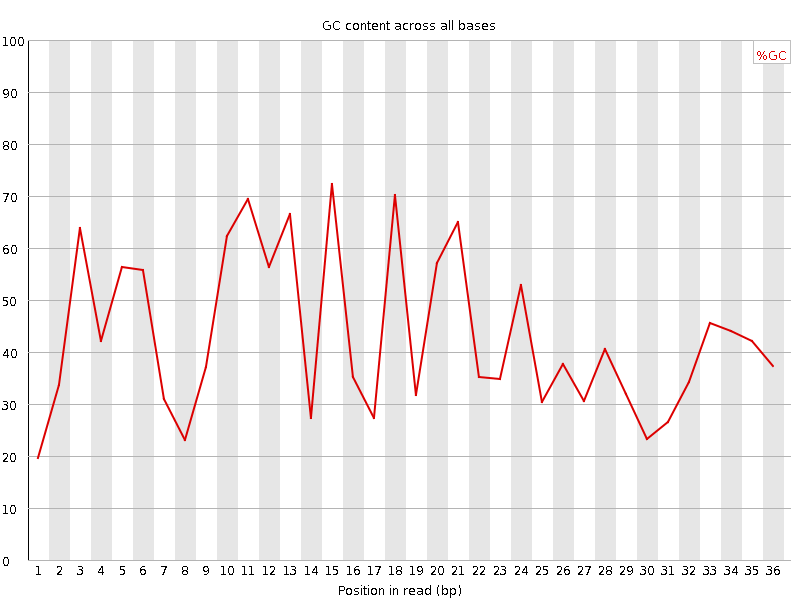
![[FAIL]](Icons/error.png) Per base GC content
Per base GC content
![[FAIL]](Icons/error.png) Per sequence GC content
Per sequence GC content
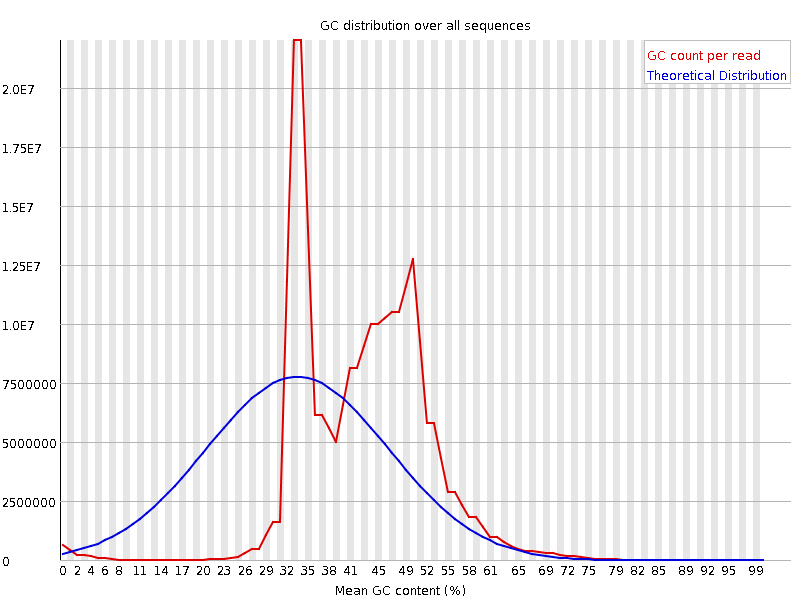
![[OK]](Icons/tick.png) Per base N content
Per base N content
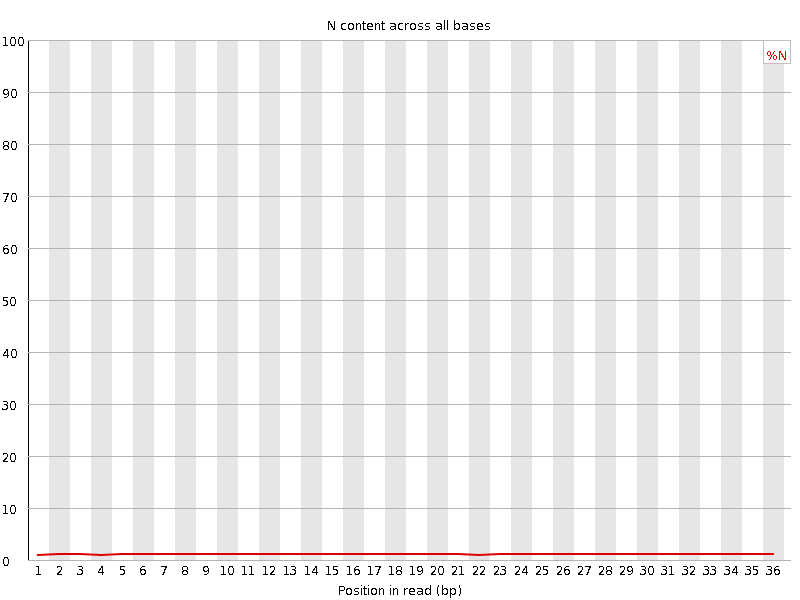
![[OK]](Icons/tick.png) Sequence Length Distribution
Sequence Length Distribution
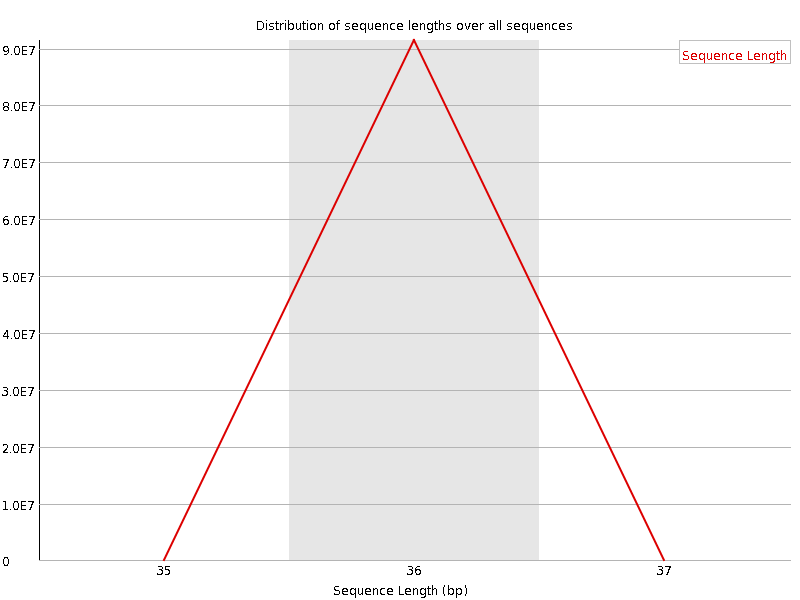
![[FAIL]](Icons/error.png) Sequence Duplication Levels
Sequence Duplication Levels
![[FAIL]](Icons/error.png) Overrepresented sequences
Overrepresented sequences
| Sequence | Count | Percentage | Possible Source |
|---|---|---|---|
| ATCTCGTATGCCGTCTTCTGCTTGAAAAAAAAAAAA | 18683288 | 20.437153413022017 | Illumina Single End Adapter 2 (95% over 24bp) |
| TAGCTTATCAGACTGGTGTTGGCATCTCGTATGCCG | 4826776 | 5.279882299212685 | No Hit |
| ATCTCGTATGCCGTCTTCTGCTTGAAAAAAAAAAAG | 1248852 | 1.366086093312878 | Illumina Single End Adapter 2 (95% over 24bp) |
| TGAGAACTGAATTCCATAGATGGATCTCGTATGCCG | 1066526 | 1.1666445157285332 | No Hit |
| TGAGATGAAGCACTGTAGCTATCTCGTATGCCGTCT | 823460 | 0.9007610624793191 | RNA PCR Primer, Index 38 (100% over 21bp) |
| GCATTGGTGGTTCAGTGGTAGAATTCTCATCTCGTA | 738498 | 0.8078233831866175 | No Hit |
| TAGCTTATCAGACTGGTGTTGGATCTCGTATGCCGT | 683306 | 0.7474503176335141 | No Hit |
| ATCTCGTATGCCGTCTTCTGCTTGAAAAAAAAAAAC | 647362 | 0.708132128978623 | Illumina Single End Adapter 2 (95% over 24bp) |
| TGAGGTAGTAGGTTGTATAGTTATCTCGTATGCCGT | 633237 | 0.6926811659597509 | No Hit |
| TTCCTATGCATATACCTCTTTATCTCGTATGCCGTC | 515911 | 0.5643413650994195 | No Hit |
| TGAGAACTGAATTCCATAGATGGTATCTCGTATGCC | 495956 | 0.5425131196451475 | No Hit |
| ATCTCGTATGCCGTCTTCTGCTTGAAAAAAAAAAAT | 489401 | 0.5353427789309028 | Illumina Single End Adapter 2 (95% over 24bp) |
| NNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNN | 442494 | 0.4840324552263908 | No Hit |
| GCATGTTTGTGGAGAACCTGGTGCTAAATCACTCGT | 371779 | 0.406679191518105 | No Hit |
| TGAGGTAGTAGATTGAATAGTTATCTCGTATGCCGT | 363491 | 0.3976131680490493 | No Hit |
| ATCTCGTATGCCGTCTTCTGCTTGAAAAAAAAAAGA | 350568 | 0.3834770409628274 | Illumina Single End Adapter 2 (95% over 24bp) |
| ATCTCGTATGCCGTCTTCTGCTTGAAAAAAAAAACA | 337614 | 0.3693070037984757 | Illumina Single End Adapter 2 (95% over 24bp) |
| TGAGATGAAGCACTGTAGCTCTATCTCGTATGCCGT | 332394 | 0.3635969841907934 | No Hit |
| ATGTTTGTGGAGAACCTGGTGCTAAATCACTCGTAT | 237819 | 0.2601438990573545 | No Hit |
| TGTAAACATCCTTGACTGGAAGCATCTCGTATGCCG | 227534 | 0.24889341023263956 | No Hit |
| TCTCGTATGCCGTCTTCTGCTTGAAAAAAAAAAAAA | 222564 | 0.2434568589969727 | Illumina Single End Adapter 2 (95% over 23bp) |
| ATTCGTATGCCGTCTTCTGCTTGAAAAAAAAAAAAA | 201640 | 0.2205686501327689 | Illumina Single End Adapter 2 (100% over 21bp) |
| TGTAAACATCCCCGACTGGAAGCATCTCGTATGCCG | 200728 | 0.21957103751165657 | No Hit |
| TGTAAACATCCTTGACTGAAAGCATCTCGTATGCCG | 193159 | 0.21129150908051728 | No Hit |
| TACCCTGTAGAACCGAATTTGTATCTCGTATGCCGT | 186726 | 0.20425462093181615 | No Hit |
| GCATTGGTGGTTCAGTGGTAGAATTCTATCTCGTAT | 182574 | 0.19971285820938378 | No Hit |
| GCATTGGTGGTTCAGTGGTAGAATTCTCGCCTGCAT | 177014 | 0.19363092161576054 | No Hit |
| GTGGAGAACCTGGTGCTAAATCACTCGTATGCCGTC | 174076 | 0.19041712130783514 | No Hit |
| TTTCCTATGCATATACCTCTTTATCTCGTATGCCGT | 173209 | 0.1894687329936856 | No Hit |
| AAGCTGCCAGCTGAAGAACTGTATCTCGTATGCCGT | 162674 | 0.17794477579695522 | No Hit |
| ATCGTATGCCGTCTTCTGCTTGAAAAAAAAAAAAAA | 159721 | 0.1747145673867089 | Illumina Single End Adapter 2 (95% over 22bp) |
| CAACTCTTAGCGGTGGATCACTCGGCATCTCGTATG | 151088 | 0.1652711575642719 | No Hit |
| TTCCTATGCATATACCTCTTTGAATCTCGTATGCCG | 145721 | 0.15940033855384456 | No Hit |
| TGAGGTAGTTGGTTGTATGGTTATCTCGTATGCCGT | 139003 | 0.1520516964610458 | No Hit |
| GCATTGGTGGTTCAGTGGTAGAATTCTCGCCATCTC | 136474 | 0.14928529040973768 | No Hit |
| TACCCTGTAGATCCGGATTTGTGATCTCGTATGCCG | 134865 | 0.14752524796744632 | No Hit |
| ATCTCGTATGCCGTCTTCTGCTTGAAAAAAAAAATA | 132671 | 0.14512528953463888 | Illumina Single End Adapter 2 (95% over 24bp) |
| TACCCTGTAGATCCGGATTTGTATCTCGTATGCCGT | 125695 | 0.13749442808192017 | No Hit |
| GCATTGGTGGTTCAGTGGTAGAATTCTCGCCTATCT | 124067 | 0.1357136020433557 | No Hit |
| TACAGTACTGTGATAACTGAAGATCTCGTATGCCGT | 121593 | 0.13300735903389094 | No Hit |
| ATAGCTCTTTGAATGGTACTGCATCTCGTATGCCGT | 121420 | 0.13281811892045625 | No Hit |
| TACCCTGTAGAACCGAATTTGTGATCTCGTATGCCG | 117733 | 0.12878500736997264 | No Hit |
| ATCTCGTATGCCGTCTTCTGCTTGAAAAAAAAAACG | 113160 | 0.1237827239090663 | Illumina Single End Adapter 2 (95% over 24bp) |
| TCCCTGAGACCCTAACTTGTGAATCTCGTATGCCGT | 111655 | 0.12213644430953338 | No Hit |
| TGAGATGAAGCACTGTAGCTCGGATCTCGTATGCCG | 107429 | 0.11751373494898448 | No Hit |
| TGTGGAGAACCTGGTGCTAAATCACTCGTATGCCGT | 101241 | 0.11074484580485844 | No Hit |
| TGAGGTAGTAGGTTGTATGGTTATCTCGTATGCCGT | 94974 | 0.10388954065517553 | No Hit |
| ATGCGCACCGCATGTTTGTGGAGAACCTGGTGCTAA | 94958 | 0.10387203867936654 | No Hit |
| TGTAAACATCCTACACTCTCAGCATCTCGTATGCCG | 93377 | 0.10214262469474093 | No Hit |
![[FAIL]](Icons/error.png) Kmer Content
Kmer Content
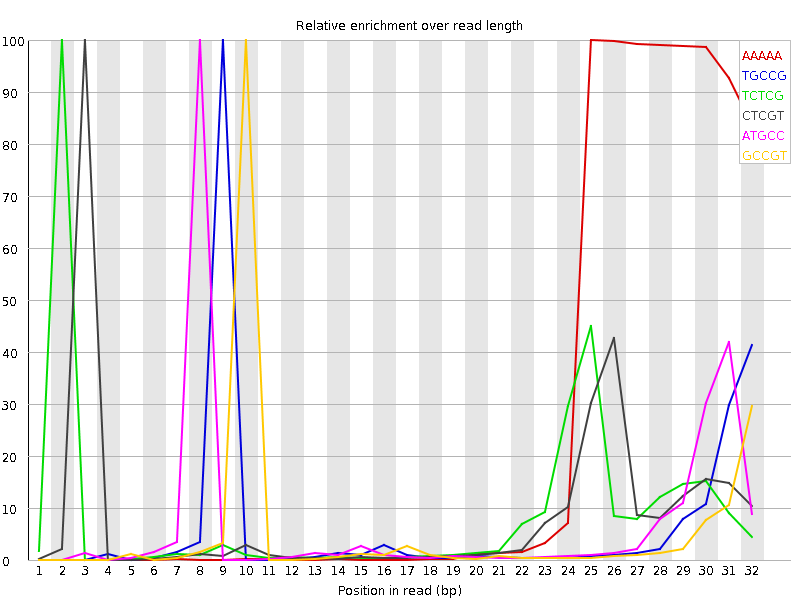
| Sequence | Count | Obs/Exp Overall | Obs/Exp Max | Max Obs/Exp Position |
|---|---|---|---|---|
| AAAAA | 211873780 | 45.484974 | 183.9745 | 25 |
| TGCCG | 53164360 | 29.122454 | 434.95868 | 9 |
| TCTCG | 69928175 | 28.93658 | 328.75354 | 2 |
| CTCGT | 69089395 | 28.589487 | 327.40634 | 3 |
| ATGCC | 55935315 | 24.456429 | 346.95898 | 8 |
| GCCGT | 42161995 | 23.09556 | 430.09125 | 10 |
| ATCTC | 69242520 | 22.870073 | 262.88614 | 1 |
| TCGTA | 68466495 | 21.617685 | 249.63495 | 4 |
| CGTAT | 64603425 | 20.397957 | 249.42311 | 5 |
| CCGTC | 34575830 | 19.812693 | 446.429 | 11 |
| GTATG | 61884905 | 18.678942 | 239.81544 | 6 |
| TATGC | 58748545 | 18.549328 | 249.46953 | 7 |
| CGTCT | 32551805 | 13.4700775 | 324.15942 | 12 |
| TCTGC | 29582780 | 12.241482 | 325.9925 | 17 |
| CTGCT | 29146500 | 12.060946 | 324.48813 | 18 |
| GCTTG | 29177655 | 11.542021 | 312.11343 | 20 |
| CTTCT | 30454535 | 9.519949 | 246.83131 | 15 |
| TCTTC | 30220390 | 9.446756 | 244.39644 | 14 |
| CTTGA | 29725020 | 9.38541 | 248.51004 | 21 |
| GTCTT | 30692150 | 9.171629 | 235.18315 | 13 |
| TTCTG | 30604145 | 9.145329 | 237.51414 | 16 |
| TGCTT | 29415640 | 8.790174 | 235.76982 | 19 |
| GAAAA | 30424350 | 8.182998 | 215.12874 | 24 |
| TGAAA | 30810005 | 7.8427954 | 202.79585 | 23 |
| TTGAA | 31007780 | 7.470297 | 191.39131 | 22 |
| CATCT | 19036795 | 6.287651 | 74.7034 | 23 |
| GCATC | 12726450 | 5.5643477 | 95.669464 | 22 |
| TGGTG | 12417860 | 4.695855 | 83.71033 | 14 |
| CTGGT | 10437360 | 4.1287837 | 86.8345 | 13 |
| GACTG | 9770020 | 4.0835595 | 93.879845 | 11 |
| GGCAT | 9119855 | 3.8118107 | 78.86247 | 21 |
| CAGAC | 8129325 | 3.755549 | 101.258415 | 9 |
| ACTGG | 8945370 | 3.7388818 | 93.832855 | 12 |
| GTTGG | 8694105 | 3.2877047 | 81.849464 | 18 |
| GATCT | 10387190 | 3.2796628 | 20.89356 | 23 |
| TAGCT | 10352215 | 3.2686198 | 71.01328 | 1 |
| TGGCA | 7501895 | 3.1355548 | 77.29888 | 20 |
| TCAGA | 8991800 | 2.999782 | 77.775986 | 8 |
| GGTGT | 7887480 | 2.9826767 | 81.90778 | 15 |
| TATCT | 12009165 | 2.8643858 | 29.500404 | 22 |
| TTATC | 11799220 | 2.8143108 | 53.49873 | 5 |
| GGATC | 6649075 | 2.779103 | 20.080975 | 22 |
| AGACT | 8245300 | 2.75074 | 74.03972 | 10 |
| ATCAG | 8222515 | 2.7431386 | 74.24159 | 7 |
| TTGGC | 6682750 | 2.643545 | 73.59967 | 19 |
| AGAAC | 7388980 | 2.604591 | 24.133842 | 3 |
| GTGGT | 6691670 | 2.5304773 | 32.839207 | 15 |
| GCTTA | 7907750 | 2.4968016 | 70.797935 | 3 |
| GGTGG | 4944200 | 2.475006 | 36.716038 | 6 |
| AGCTT | 7792030 | 2.460264 | 70.94616 | 2 |
| TGTTG | 8281930 | 2.3658495 | 60.94535 | 17 |
| GGTAG | 5665080 | 2.2635283 | 28.141087 | 17 |
| GTGTT | 7793660 | 2.2263684 | 60.606453 | 16 |
| GAGAA | 5946365 | 2.0037477 | 22.750805 | 2 |
| CTTAT | 8390025 | 2.001161 | 53.533928 | 4 |
| TATCA | 7841445 | 1.9761814 | 56.063396 | 6 |
| GTAGA | 6149120 | 1.9610672 | 22.460672 | 18 |
| TGAGA | 6063300 | 1.9336977 | 42.74986 | 1 |
| CCTGG | 3254720 | 1.7828755 | 9.10794 | 17 |
| AATCT | 6931780 | 1.74693 | 6.900384 | 22 |
| CTGAA | 5195705 | 1.7333552 | 21.175228 | 7 |
| GTGGA | 4327885 | 1.7292414 | 8.884996 | 13 |
| AGTGG | 4316215 | 1.7245786 | 34.97893 | 14 |
| GAAGC | 3820240 | 1.6871204 | 30.380623 | 7 |
| CAGTG | 3899800 | 1.6299932 | 28.355988 | 13 |
| GAACC | 3524875 | 1.6284058 | 8.698848 | 10 |
| AGATG | 5074040 | 1.6182047 | 21.300575 | 3 |
| GGAGA | 3821840 | 1.6134828 | 6.7581716 | 11 |
| ACTGA | 4821965 | 1.6086706 | 21.552034 | 6 |
| TGTAG | 5329010 | 1.6084741 | 20.45767 | 14 |
| CACTC | 3497755 | 1.5997787 | 9.178919 | 19 |
| TGGAT | 5284465 | 1.595029 | 12.473927 | 21 |
| ACTCG | 3637760 | 1.5905268 | 8.895717 | 20 |
| CTGTA | 4941460 | 1.560222 | 21.489029 | 13 |
| GAATT | 6371690 | 1.5350478 | 16.10829 | 21 |
| AAGCA | 4336930 | 1.5287535 | 23.919006 | 8 |
| TGAAG | 4701285 | 1.4993261 | 21.854595 | 6 |
| TGGTT | 5233470 | 1.4950142 | 18.829115 | 8 |
| GAACT | 4467255 | 1.4903347 | 22.05235 | 4 |
| GATGA | 4566000 | 1.4561814 | 22.43505 | 4 |
| TGGAG | 3632620 | 1.4514425 | 6.101409 | 10 |
| CACTG | 3294145 | 1.440289 | 30.314342 | 11 |
| CTCGG | 2611415 | 1.4304848 | 10.49173 | 21 |
| GGTTC | 3575220 | 1.4142764 | 26.138649 | 9 |
| TGGTA | 4676085 | 1.4113996 | 20.823143 | 16 |
| AACTG | 4217625 | 1.4070548 | 21.187584 | 5 |
| AGCAC | 3020435 | 1.3953669 | 30.891474 | 9 |
| TGAGG | 3483920 | 1.3920282 | 27.089521 | 1 |
| TGTGG | 3670155 | 1.3878813 | 6.220727 | 8 |
| ACTGT | 4389035 | 1.3857987 | 22.118832 | 12 |
| TGAAT | 5646825 | 1.3604153 | 15.305051 | 8 |
| GCACT | 3086350 | 1.3494356 | 29.83928 | 10 |
| GGTGC | 2524680 | 1.3220569 | 7.941355 | 20 |
| GATGG | 3308455 | 1.3219199 | 24.46532 | 19 |
| CTATC | 3992180 | 1.3185749 | 10.630307 | 19 |
| ACCTG | 2980235 | 1.3030392 | 8.780358 | 16 |
| TAGAA | 5072995 | 1.2913488 | 17.27299 | 19 |
| ATCAC | 3617235 | 1.2623603 | 6.834846 | 17 |
| GTAGC | 2998170 | 1.2531402 | 27.053118 | 15 |
| AATTC | 4937785 | 1.2444084 | 16.22911 | 22 |
| CCCTG | 2158620 | 1.2369356 | 14.5686655 | 3 |
| AAAAG | 4574735 | 1.2304304 | 14.948413 | 32 |
| CTCTT | 3881740 | 1.2134143 | 9.509876 | 16 |
| AGCAT | 3592665 | 1.19856 | 10.8769 | 21 |
| GTATC | 3770550 | 1.1905174 | 8.483774 | 21 |
| GAGGT | 2969540 | 1.1865036 | 24.26504 | 2 |
| GGAAG | 2799545 | 1.1818961 | 10.052793 | 18 |
| ATGAA | 4631395 | 1.1789379 | 17.12176 | 5 |
| TCATC | 3558895 | 1.1754655 | 12.231256 | 27 |
| GAATC | 3498530 | 1.1671555 | 5.0382795 | 22 |
| ACATC | 3342705 | 1.1665534 | 14.64853 | 6 |
| AGAAT | 4553905 | 1.1592127 | 17.490072 | 20 |
| TCAGT | 3669860 | 1.1587256 | 24.234879 | 12 |
| GTGCT | 2903870 | 1.1487054 | 5.8493004 | 21 |
| AGCGG | 2058690 | 1.1390603 | 11.916424 | 9 |
| CATTG | 3599575 | 1.1365337 | 24.008745 | 2 |
| GGTTG | 3004820 | 1.1362827 | 16.761532 | 11 |
| GAGAT | 3554400 | 1.1335634 | 22.073679 | 2 |
| CTGGA | 2663105 | 1.1130936 | 9.903884 | 16 |
| AGATC | 3298600 | 1.1004562 | 5.0345893 | 21 |
| GTAGG | 2751100 | 1.0992241 | 14.688561 | 8 |
| TTGGT | 3813425 | 1.0893584 | 19.556427 | 4 |
| AGCTC | 2487450 | 1.0875803 | 14.387422 | 17 |
| TCACT | 3261395 | 1.0772041 | 6.5989223 | 18 |
| CCGGC | 1409075 | 1.0688515 | 8.021632 | 11 |
| TTCAG | 3343770 | 1.0557656 | 20.569574 | 11 |
| TGCAT | 3317960 | 1.0476164 | 10.130285 | 7 |
| TCGGC | 1911705 | 1.0471966 | 9.14236 | 22 |
| AAACA | 3713025 | 1.0446782 | 11.682692 | 4 |
| TTCTC | 3332810 | 1.0418212 | 17.333704 | 24 |
| CGGTG | 1987120 | 1.0405618 | 10.894863 | 11 |
| AAAAC | 3693185 | 1.039096 | 7.68763 | 32 |
| GCATT | 3247100 | 1.0252429 | 23.567944 | 1 |
| GTTCA | 3246320 | 1.0249966 | 20.664406 | 10 |
| CGGCT | 1850040 | 1.0134176 | 5.794045 | 12 |
| TAGAT | 4197045 | 1.0111389 | 14.868728 | 17 |
| TGGGG | 2019020 | 1.0106968 | 5.6137686 | 14 |
| TGGAA | 3133840 | 0.9994391 | 7.514404 | 17 |
| ACCTC | 2178745 | 0.9964991 | 13.182308 | 14 |
| GTAGT | 3269160 | 0.98674226 | 18.190996 | 5 |
| ATTGG | 3250230 | 0.98102856 | 21.032364 | 3 |
| TCCAT | 2904180 | 0.9592198 | 21.058271 | 13 |
| GTCGG | 1812720 | 0.94923675 | 6.7411394 | 14 |
| GCGGT | 1798475 | 0.9417773 | 12.178494 | 10 |
| AACCT | 2695705 | 0.94076014 | 6.2255955 | 15 |
| GTGGG | 1873500 | 0.93785125 | 5.0084767 | 18 |
| CCTCT | 2160070 | 0.93503183 | 13.444003 | 15 |
| ATGCA | 2790070 | 0.93080384 | 10.451269 | 6 |
| TTGGA | 3028640 | 0.9141453 | 9.155383 | 19 |
| ATTCC | 2760550 | 0.91178036 | 20.607525 | 11 |
| GGGGT | 1815890 | 0.9090124 | 6.2174444 | 15 |
| GGGCA | 1640445 | 0.9076479 | 5.016383 | 17 |
| CCTGT | 2188645 | 0.90567076 | 10.523105 | 4 |
| AGTAG | 2835580 | 0.90431863 | 17.649046 | 7 |
| AAATC | 3385655 | 0.90154076 | 5.246543 | 26 |
| GATCA | 2672975 | 0.8917394 | 6.367653 | 16 |
| TGTAT | 3873250 | 0.8831425 | 10.934547 | 14 |
| AGGTA | 2752325 | 0.87776697 | 18.958878 | 3 |
| AACAT | 3291645 | 0.8765075 | 11.169159 | 5 |
| CCTAT | 2651180 | 0.8756568 | 10.462373 | 3 |
| ATGGA | 2738230 | 0.8732718 | 13.117923 | 20 |
| AACTC | 2498040 | 0.8717781 | 7.3394933 | 2 |
| ACTCT | 2638560 | 0.87148833 | 7.155 | 3 |
| TTCCA | 2637695 | 0.87120265 | 20.899742 | 12 |
| GGCCG | 1198045 | 0.868746 | 10.374494 | 9 |
| GCTAA | 2600645 | 0.8676092 | 5.2625365 | 23 |
| CCATA | 2457330 | 0.8575709 | 21.368065 | 14 |
| GCCGG | 1152865 | 0.83598435 | 8.195996 | 10 |
| CATAG | 2475750 | 0.8259426 | 20.601702 | 15 |
| CTCAT | 2494865 | 0.8240274 | 11.510118 | 26 |
| CTGGG | 1568420 | 0.8213082 | 5.1486816 | 13 |
| CGGCA | 1417630 | 0.8205068 | 5.138934 | 23 |
| CATCC | 1792350 | 0.8197725 | 19.00553 | 7 |
| TTTGT | 3788000 | 0.81743497 | 5.7523947 | 18 |
| CTCAG | 1857070 | 0.8119611 | 6.222414 | 7 |
| GTTGT | 2837695 | 0.8106274 | 12.583424 | 12 |
| AAAGA | 2978970 | 0.8012301 | 5.129333 | 32 |
| TCTCA | 2413380 | 0.7971138 | 11.357652 | 25 |
| GCATG | 1906090 | 0.7966854 | 6.994048 | 1 |
| TTGTA | 3484665 | 0.7945409 | 10.697486 | 13 |
| ATTCT | 3309550 | 0.7893828 | 14.794092 | 23 |
| ACCCT | 1677880 | 0.76741695 | 11.933029 | 2 |
| TAGTA | 3174035 | 0.7646785 | 12.4065275 | 6 |
| CTATG | 2412415 | 0.76169854 | 10.071534 | 4 |
| TGATG | 2494850 | 0.75302947 | 5.0807652 | 7 |
| GCATA | 2252910 | 0.75160027 | 10.227113 | 8 |
| GCTCT | 1797150 | 0.74366844 | 8.755847 | 18 |
| TCCTA | 2247490 | 0.7423221 | 10.281425 | 2 |
| ATAGA | 2866505 | 0.72967887 | 15.595601 | 16 |
| TAGCG | 1740525 | 0.7274844 | 8.936808 | 8 |
| CTCGC | 1248565 | 0.71545446 | 11.779587 | 26 |
| ATACC | 2034035 | 0.70984733 | 10.600174 | 12 |
| TGACT | 2231090 | 0.7044468 | 6.864544 | 13 |
| GCTAT | 2213150 | 0.6987824 | 10.053887 | 18 |
| GCCGA | 1204635 | 0.6972279 | 7.7619314 | 10 |
| ATGGT | 2283605 | 0.68926865 | 5.910172 | 20 |
| CGCCT | 1201730 | 0.6886171 | 7.6975894 | 28 |
| TGTAA | 2836225 | 0.6832944 | 10.3201275 | 1 |
| GATTG | 2253580 | 0.6802061 | 5.4567 | 11 |
| TTCCT | 2167820 | 0.6776507 | 10.094077 | 1 |
| AGCTA | 2021895 | 0.6745306 | 10.507415 | 17 |
| GCTGT | 1704275 | 0.6741727 | 5.426802 | 11 |
| CTGCC | 1170595 | 0.670776 | 5.347458 | 4 |
| TTGAC | 2118110 | 0.66877437 | 6.777485 | 12 |
| GGTCG | 1275615 | 0.6679799 | 5.4781756 | 4 |
| CTGTC | 1613335 | 0.667605 | 5.410573 | 12 |
| GTGGC | 1263270 | 0.6615155 | 6.955556 | 7 |
| CATAT | 2605135 | 0.6565397 | 7.787893 | 9 |
| AGGGC | 1185900 | 0.6561511 | 5.0109215 | 16 |
| GGTAT | 2125905 | 0.6416695 | 6.081341 | 22 |
| AGGTT | 2106780 | 0.6358969 | 10.738751 | 10 |
| GTTAT | 2784455 | 0.63488555 | 13.086024 | 20 |
| GCTCA | 1432420 | 0.6262927 | 6.6216245 | 10 |
| GACGA | 1416775 | 0.62568575 | 5.8486786 | 16 |
| AAAAT | 3048220 | 0.6193342 | 5.110267 | 32 |
| GGTTA | 2035085 | 0.61425704 | 6.373385 | 17 |
| GACCT | 1400545 | 0.6123561 | 5.8876805 | 4 |
| TTAGC | 1912185 | 0.6037554 | 7.519338 | 7 |
| CAACT | 1728390 | 0.6031819 | 7.3632736 | 1 |
| CCCCG | 759415 | 0.60259557 | 8.926954 | 10 |
| CCGGA | 1034375 | 0.59868354 | 6.6375127 | 13 |
| CCGAA | 1289280 | 0.59561574 | 6.546056 | 13 |
| TCTTT | 2625990 | 0.5927886 | 7.1614394 | 17 |
| GCTGC | 1081560 | 0.5924586 | 5.50778 | 3 |
| AGACG | 1325160 | 0.5852262 | 5.508287 | 15 |
| TCGCC | 1015150 | 0.5817027 | 10.799823 | 27 |
| CTTAG | 1825420 | 0.5763601 | 6.87648 | 6 |
| CATGT | 1825040 | 0.5762401 | 5.199512 | 2 |
| GTAAA | 2245950 | 0.57171446 | 10.311877 | 2 |
| ATTGA | 2371355 | 0.5712993 | 5.870613 | 12 |
| TACCT | 1715635 | 0.566656 | 9.262163 | 13 |
| TCCCT | 1308025 | 0.56620616 | 7.016861 | 1 |
| TAGTT | 2482455 | 0.5660263 | 10.409795 | 18 |
| CCGAC | 932155 | 0.56437916 | 6.6526194 | 12 |
| TAGGT | 1850835 | 0.5586441 | 10.304472 | 9 |
| ATCCT | 1690080 | 0.55821544 | 10.065506 | 8 |
| CCTCA | 1213985 | 0.5552439 | 5.9775286 | 6 |
| AACCG | 1195825 | 0.5524418 | 6.6428246 | 11 |
| ATAGT | 2284165 | 0.55029374 | 11.347147 | 17 |
| TCTTA | 2298355 | 0.54819596 | 5.105699 | 5 |
| GCCAG | 934150 | 0.5406745 | 5.512122 | 6 |
| TCCGG | 980550 | 0.53712714 | 6.3197575 | 12 |
| CCTTG | 1290320 | 0.53394 | 8.736389 | 10 |
| ATCCC | 1167075 | 0.53378856 | 6.6512666 | 8 |
| TACCC | 1117165 | 0.5109611 | 11.441719 | 1 |
| GATCC | 1156505 | 0.5056552 | 5.7982984 | 10 |
| TAAAC | 1890200 | 0.5033272 | 10.672844 | 3 |
| CGCTG | 915765 | 0.5016391 | 5.0078883 | 10 |
| CCAGC | 826385 | 0.50034004 | 5.5954566 | 7 |
| TCCCC | 820390 | 0.49176174 | 7.0307446 | 9 |
| GTATA | 2039225 | 0.49128363 | 7.8957167 | 15 |
| TATAG | 1947120 | 0.46909398 | 8.026111 | 16 |
| TATAC | 1843305 | 0.4645451 | 7.7462835 | 11 |
| CTTTA | 1936945 | 0.46199366 | 5.568368 | 18 |
| CGACC | 751920 | 0.45525473 | 8.024692 | 3 |
| TGGCC | 827115 | 0.45307827 | 5.941936 | 8 |
| TCCTT | 1443425 | 0.45120808 | 6.694616 | 9 |
| AGTTA | 1817920 | 0.43796757 | 10.655343 | 19 |
| ACGAC | 946290 | 0.4371628 | 6.2496676 | 2 |
| CCCGA | 721115 | 0.43660367 | 6.742968 | 11 |
| ACCGA | 944750 | 0.43645135 | 6.5840654 | 12 |
| ATTTG | 1855380 | 0.42304644 | 5.9763813 | 17 |
| ATATA | 1942665 | 0.3735637 | 5.934087 | 10 |
| CCGAG | 592720 | 0.34305903 | 5.758648 | 11 |