Input data and parameters
QualiMap command line
| qualimap bamqc -bam SRR7062655.recal.bam -c -nw 400 -hm 3 -sd |
Alignment
| Command line: | bwa mem -K 100000000 -R @RG\tID:1\tPU:1\tSM:SRR7062655\tLB:SRR7062655\tPL:illumina -t 10 -M Gallus_gallus.Gallus_gallus-5.0.dna.toplevel_chr25-26.fa SRR7062655_R1.fastq.gz SRR7062655_R2.fastq.gz |
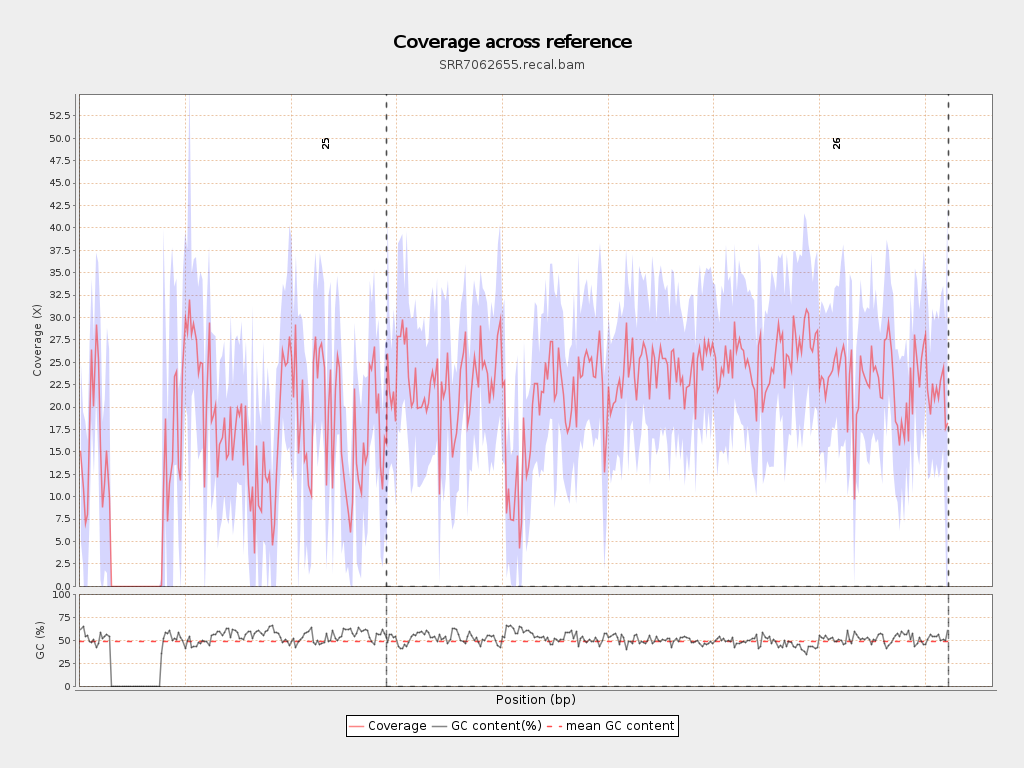
| Draw chromosome limits: | yes |
| Analyze overlapping paired-end reads: | no |
| Program: | bwa (0.7.17-r1188) |
| Analysis date: | Mon Oct 25 12:13:11 UTC 2021 |
| Size of a homopolymer: | 3 |
| Skip duplicate alignments: | yes (only flagged) |
| Number of windows: | 400 |
| BAM file: | SRR7062655.recal.bam |
Summary
Globals
| Reference size | 8,220,070 |
| Number of reads | 1,921,198 |
| Mapped reads | 1,919,576 / 99.92% |
| Unmapped reads | 1,622 / 0.08% |
| Mapped paired reads | 1,919,576 / 99.92% |
| Mapped reads, first in pair | 959,862 / 49.96% |
| Mapped reads, second in pair | 959,714 / 49.95% |
| Mapped reads, both in pair | 1,917,954 / 99.83% |
| Mapped reads, singletons | 1,622 / 0.08% |
| Secondary alignments | 721 |
| Read min/max/mean length | 30 / 90 / 90.02 |
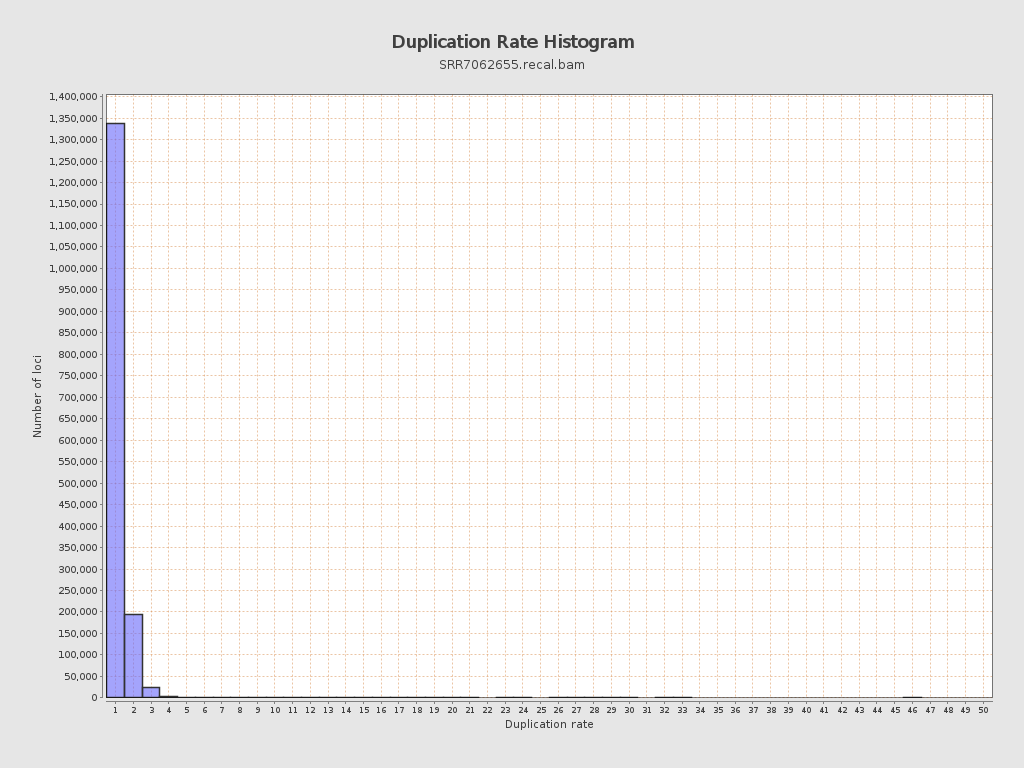
| Duplicated reads (flagged) | 86,798 / 4.52% |
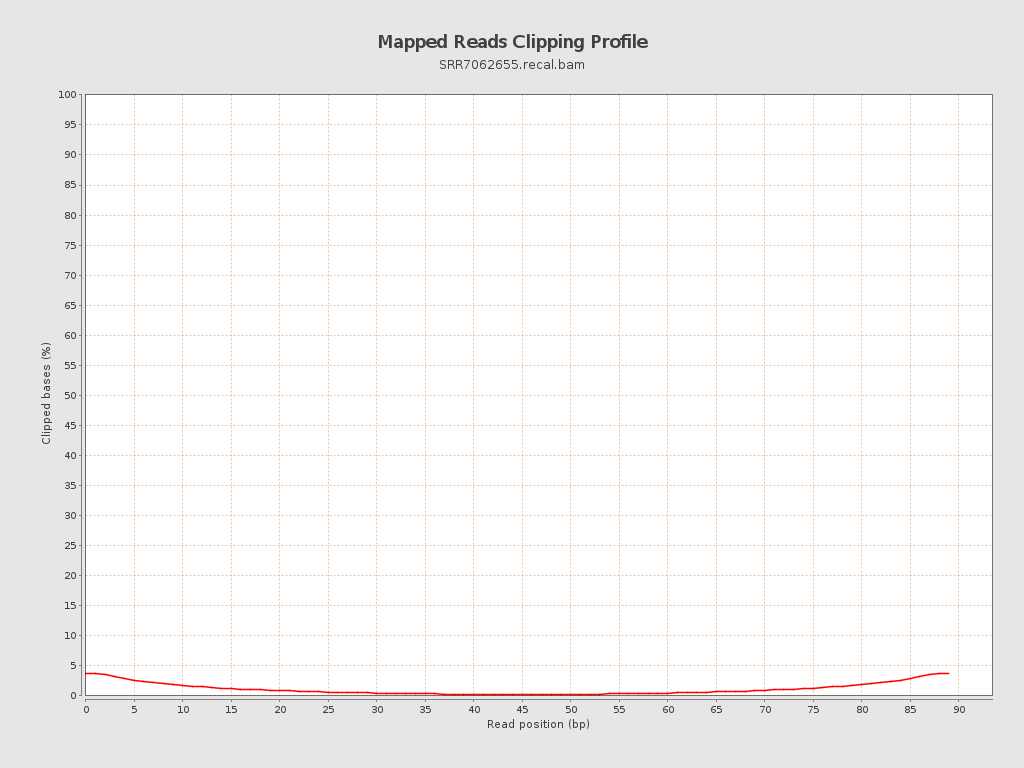
| Clipped reads | 54,147 / 2.82% |
| Duplicated reads skipped: | 86,798 / 4.52% |
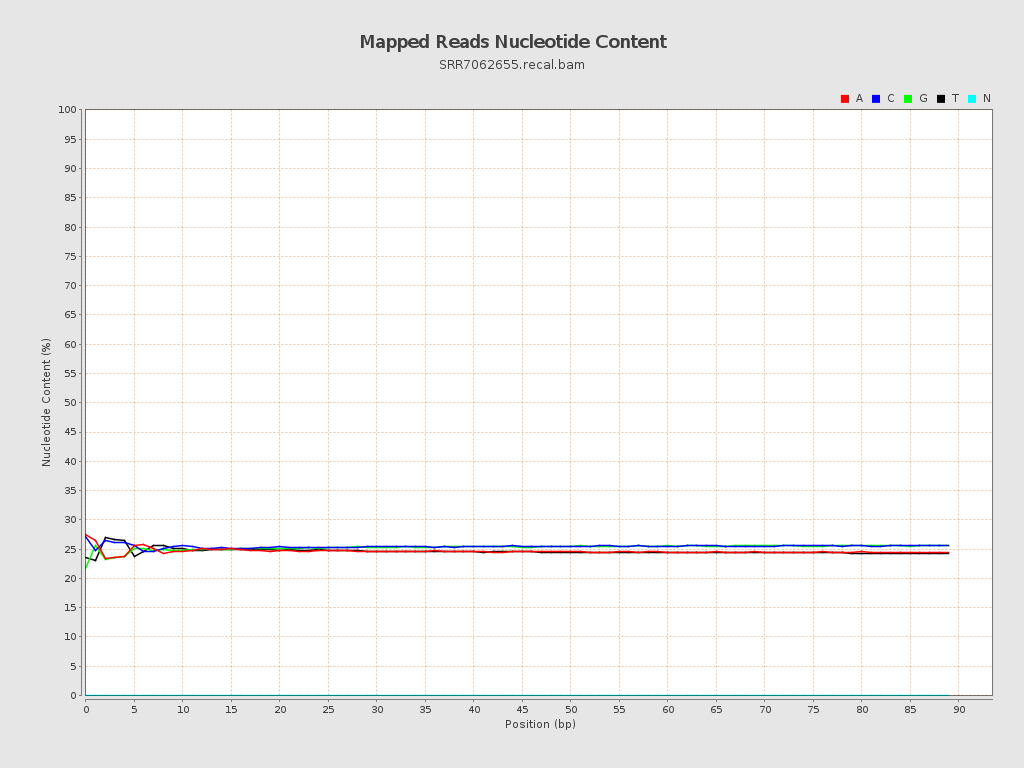
ACGT Content
| Number/percentage of A's | 40,414,310 / 24.63% |
| Number/percentage of C's | 41,798,019 / 25.47% |
| Number/percentage of T's | 40,389,320 / 24.61% |
| Number/percentage of G's | 41,491,642 / 25.28% |
| Number/percentage of N's | 22,986 / 0.01% |
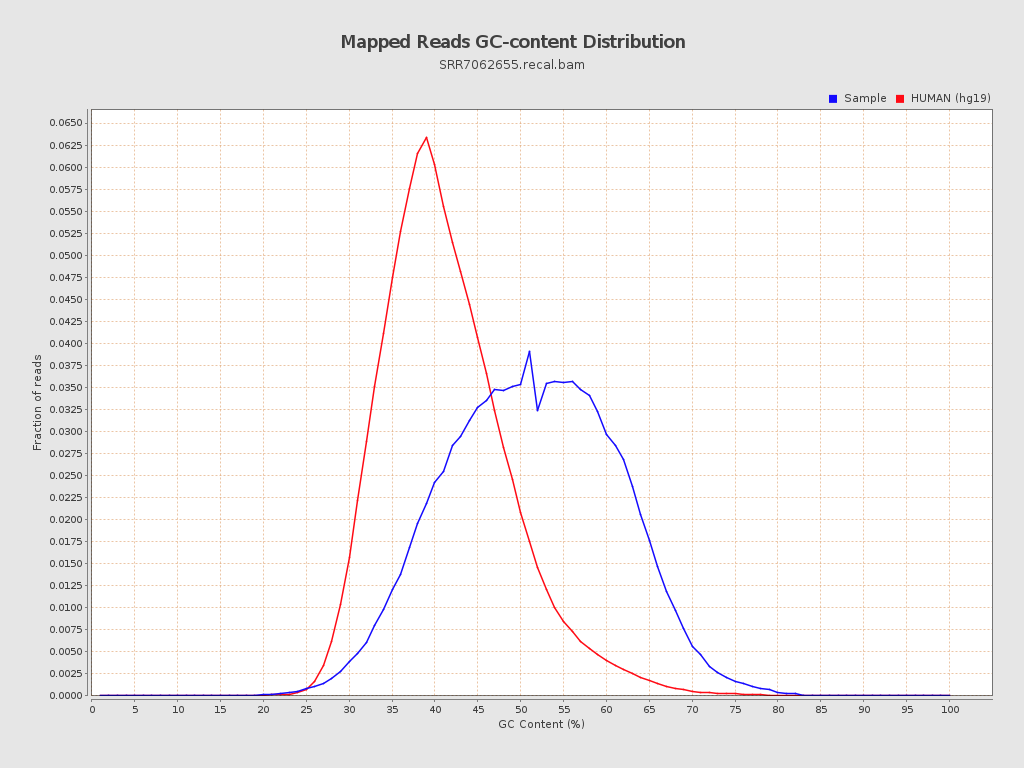
| GC Percentage | 50.75% |
Coverage
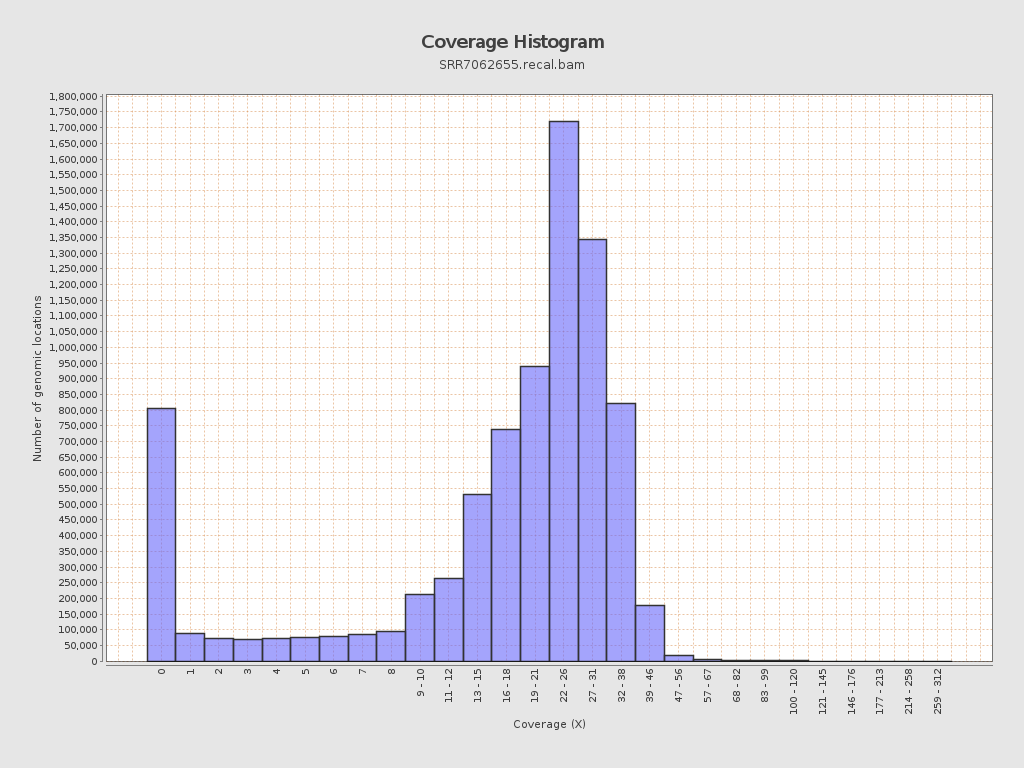
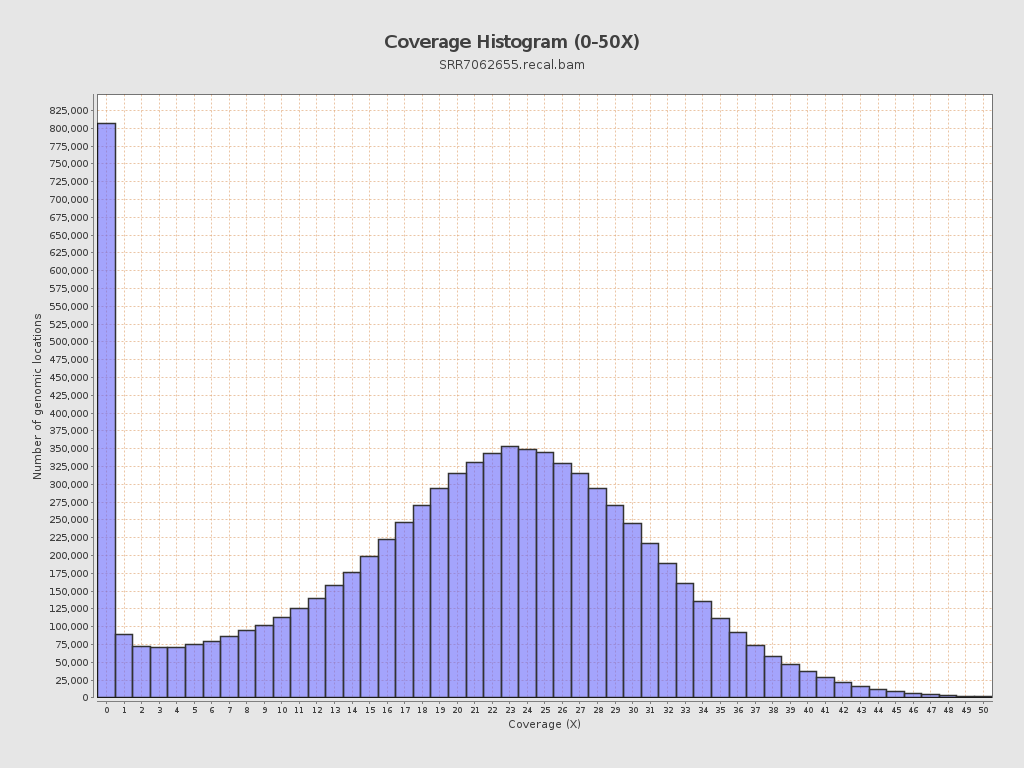
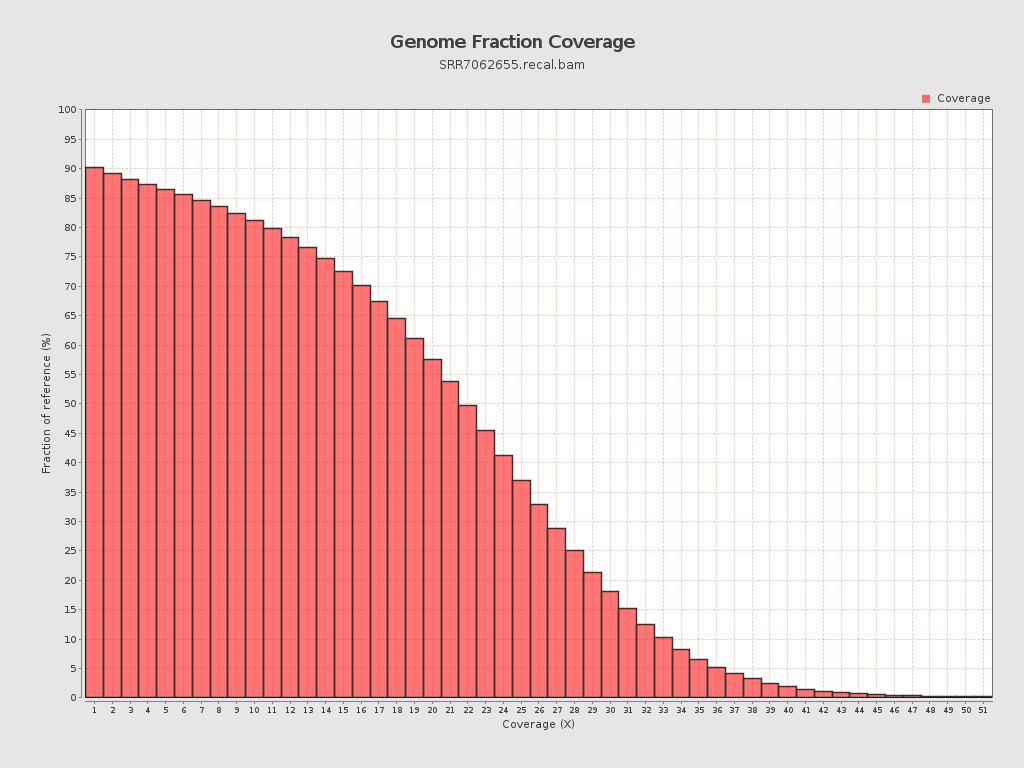
| Mean | 19.9781 |
| Standard Deviation | 11.0986 |
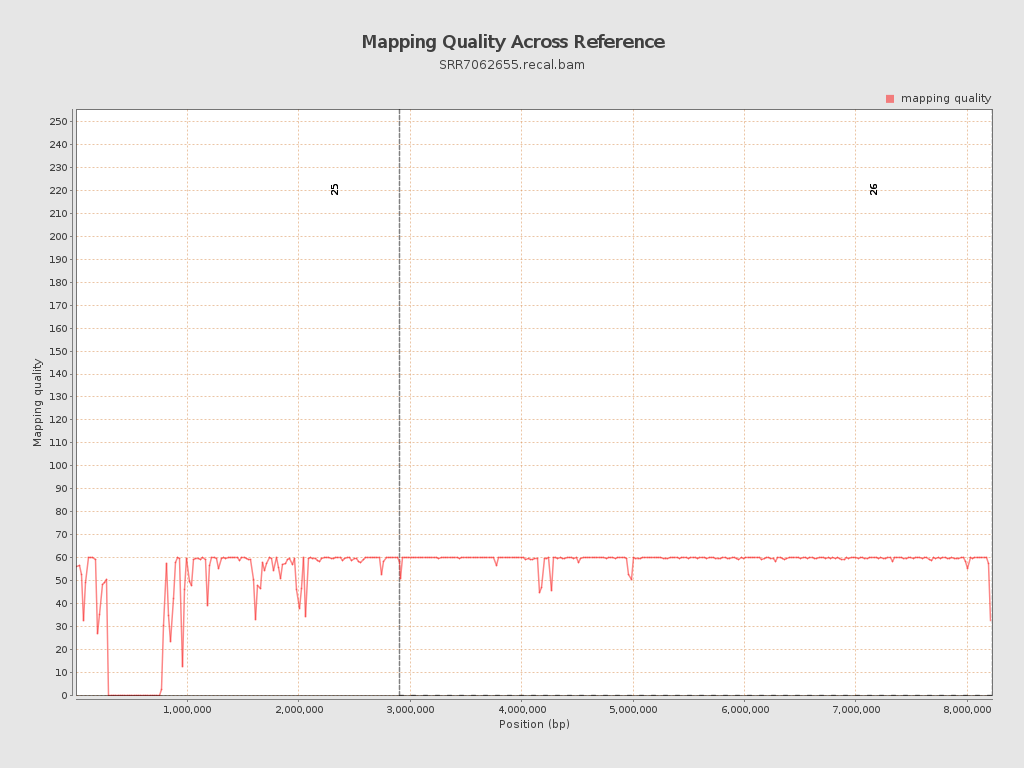
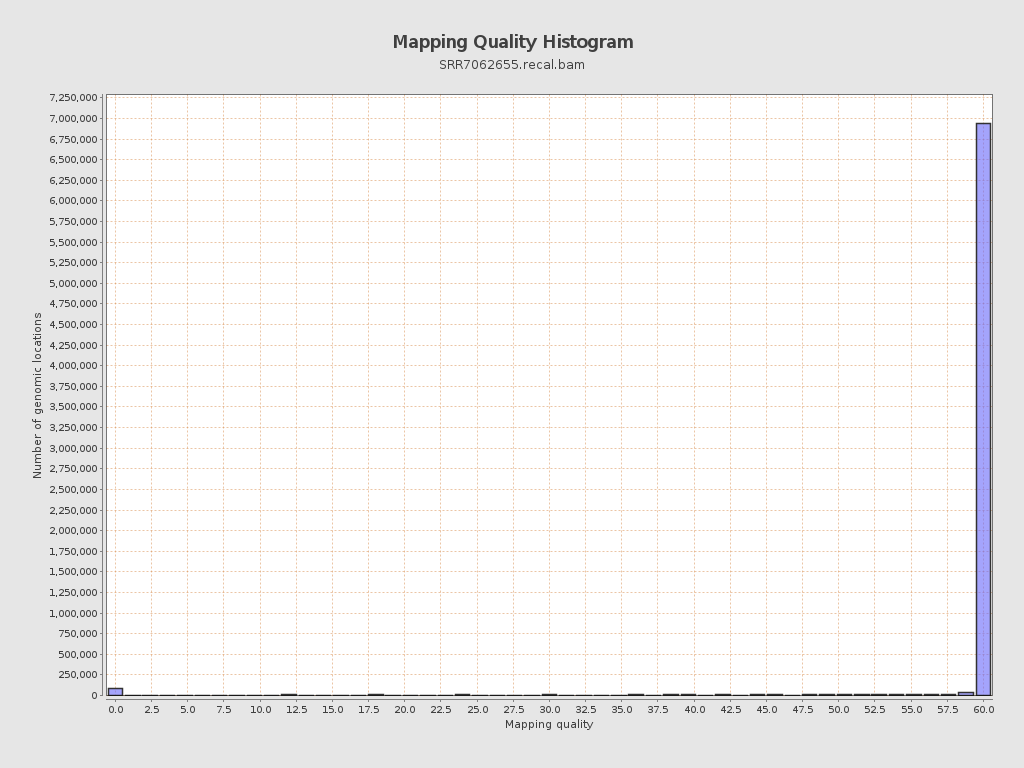
Mapping Quality
| Mean Mapping Quality | 54.63 |
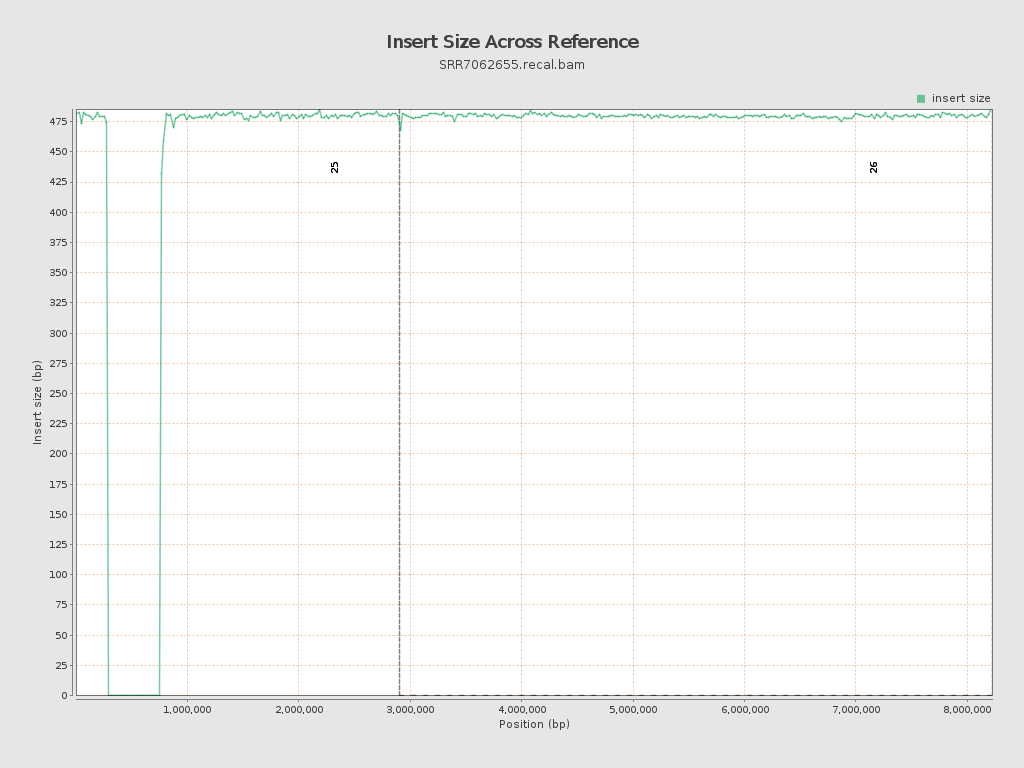
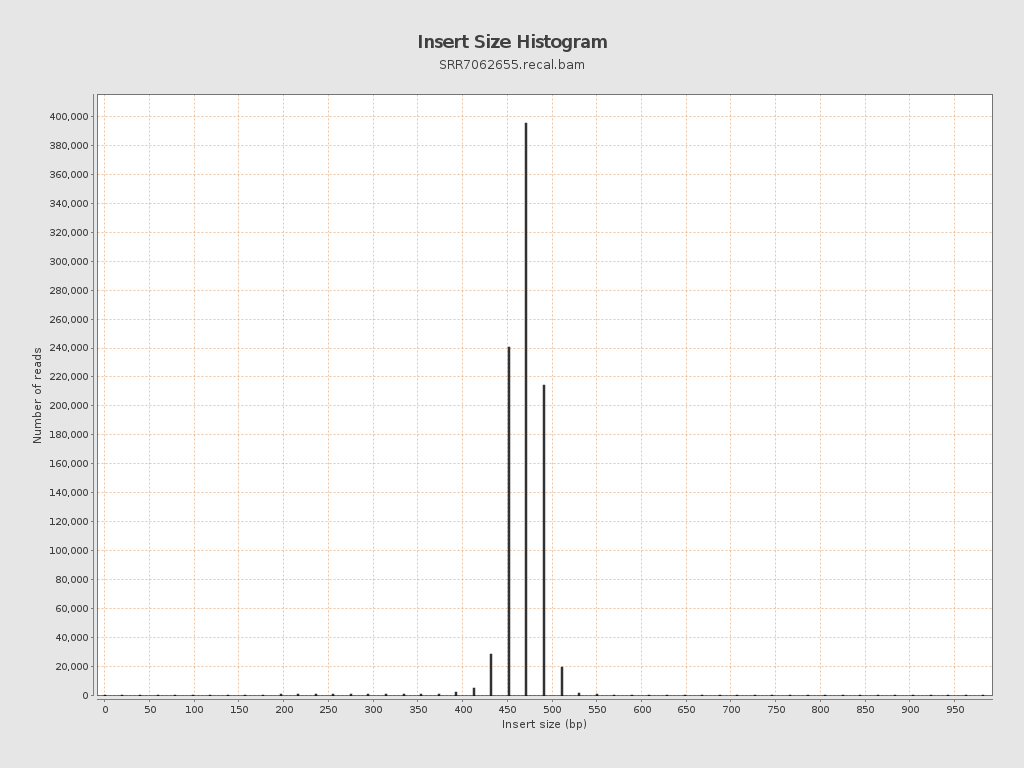
Insert size
| Mean | 628.34 |
| Standard Deviation | 14,810.5 |
| P25/Median/P75 | 469 / 480 / 491 |
Mismatches and indels
| General error rate | 0.98% |
| Mismatches | 1,537,639 |
| Insertions | 31,672 |
| Mapped reads with at least one insertion | 1.61% |
| Deletions | 36,571 |
| Mapped reads with at least one deletion | 1.84% |
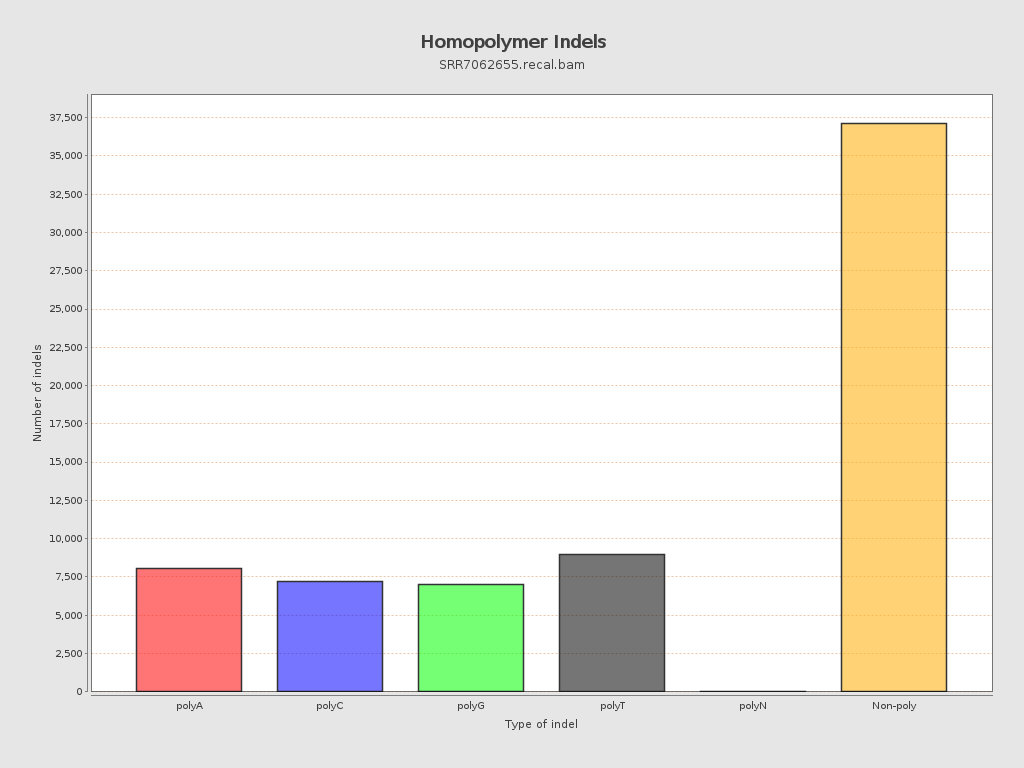
| Homopolymer indels | 45.59% |
Chromosome stats
| Name | Length | Mapped bases | Mean coverage | Standard deviation |
| 25 | 2906300 | 42585731 | 14.6529 | 12.7971 |
| 26 | 5313770 | 121635558 | 22.8906 | 8.7749 |